
Mar:Z And Be.Yond .. De Dis.Ney .. Her:Moz., Ez:Pe:KU.la.cion So.Bre. LA Vi:Da .en El . PLane:Ta ((Mar:Te)) GraN,Dio:Zaz Y es.Tram.Boti:KAz Cri-A-tur/As

Mar:Z And Be.Yond .. De Dis.Ney .. Her:Moz., Ez:Pe:KU.la.cion So.Bre. LA Vi:Da .en El . PLane:Ta ((Mar:Te)) GraN,Dio:Zaz Y es.Tram.Boti:KAz Cri-A-tur/As

A powerful gene-editing technology is the biggest game changer to hit biology since PCR. But with its huge potential come pressing concerns.
Three years ago, Bruce Conklin came across a method that made him change the course of his lab.
Conklin, a geneticist at the Gladstone Institutes in San Francisco, California, had been trying to work out how variations in DNA affect various human diseases, but his tools were cumbersome. When he worked with cells from patients, it was hard to know which sequences were important for disease and which were just background noise. And engineering a mutation into cells was expensive and laborious work. “It was a student’s entire thesis to change one gene,” he says.
Nature special:CRISPR
Then, in 2012, he read about a newly published technique1 called CRISPR that would allow researchers to quickly change the DNA of nearly any organism — including humans. Soon after, Conklin abandoned his previous approach to modelling disease and adopted this new one. His lab is now feverishly altering genes associated with various heart conditions. “CRISPR is turning everything on its head,” he says.
The sentiment is widely shared: CRISPR is causing a major upheaval in biomedical research. Unlike other gene-editing methods, it is cheap, quick and easy to use, and it has swept through labs around the world as a result. Researchers hope to use it to adjust human genes to eliminate diseases, create hardier plants, wipe out pathogens and much more besides. “I’ve seen two huge developments since I’ve been in science: CRISPR and PCR,” says John Schimenti, a geneticist at Cornell University in Ithaca, New York. Like PCR, the gene-amplification method that revolutionized genetic engineering after its invention in 1985, “CRISPR is impacting the life sciences in so many ways,” he says.
Reporter Kerri Smith investigates the meteoric rise of CRISPR

But although CRISPR has much to offer, some scientists are worried that the field’s breakneck pace leaves little time for addressing the ethical and safety concerns such experiments can raise. The problem was thrust into the spotlight in April, when news broke that scientists had used CRISPR to engineer human embryos (see Nature 520, 593–595; 2015). The embryos they used were unable to result in a live birth, but the report2 has generated heated debate over whether and how CRISPR should be used to make heritable changes to the human genome. And there are other concerns. Some scientists want to see more studies that probe whether the technique generates stray and potentially risky genome edits; others worry that edited organisms could disrupt entire ecosystems.
“This power is so easily accessible by labs — you don’t need a very expensive piece of equipment and people don’t need to get many years of training to do this,” says Stanley Qi, a systems biologist at Stanford University in California. “We should think carefully about how we are going to use that power.”
Biologists have long been able to edit genomes with molecular tools. About ten years ago, they became excited by enzymes called zinc finger nucleases that promised to do this accurately and efficiently. But zinc fingers, which cost US$5,000 or more to order, were not widely adopted because they are difficult to engineer and expensive, says James Haber, a molecular biologist at Brandeis University in Waltham, Massachusetts. CRISPR works differently: it relies on an enzyme called Cas9 that uses a guide RNA molecule to home in on its target DNA, then edits the DNA to disrupt genes or insert desired sequences. Researchers often need to order only the RNA fragment; the other components can be bought off the shelf. Total cost: as little as $30. “That effectively democratized the technology so that everyone is using it,” says Haber. “It’s a huge revolution.”
CRISPR methodology is quickly eclipsing zinc finger nucleases and other editing tools (see ‘The rise of CRISPR’). For some, that means abandoning techniques they had taken years to perfect. “I’m depressed,” says Bill Skarnes, a geneticist at the Wellcome Trust Sanger Institute in Hinxton, UK, “but I’m also excited.” Skarnes had spent much of his career using a technology introduced in the mid-1980s: inserting DNA into embryonic stem cells and then using those cells to generate genetically modified mice. The technique became a laboratory workhorse, but it was also time-consuming and costly. CRISPR takes a fraction of the time, and Skarnes adopted the technique two years ago.
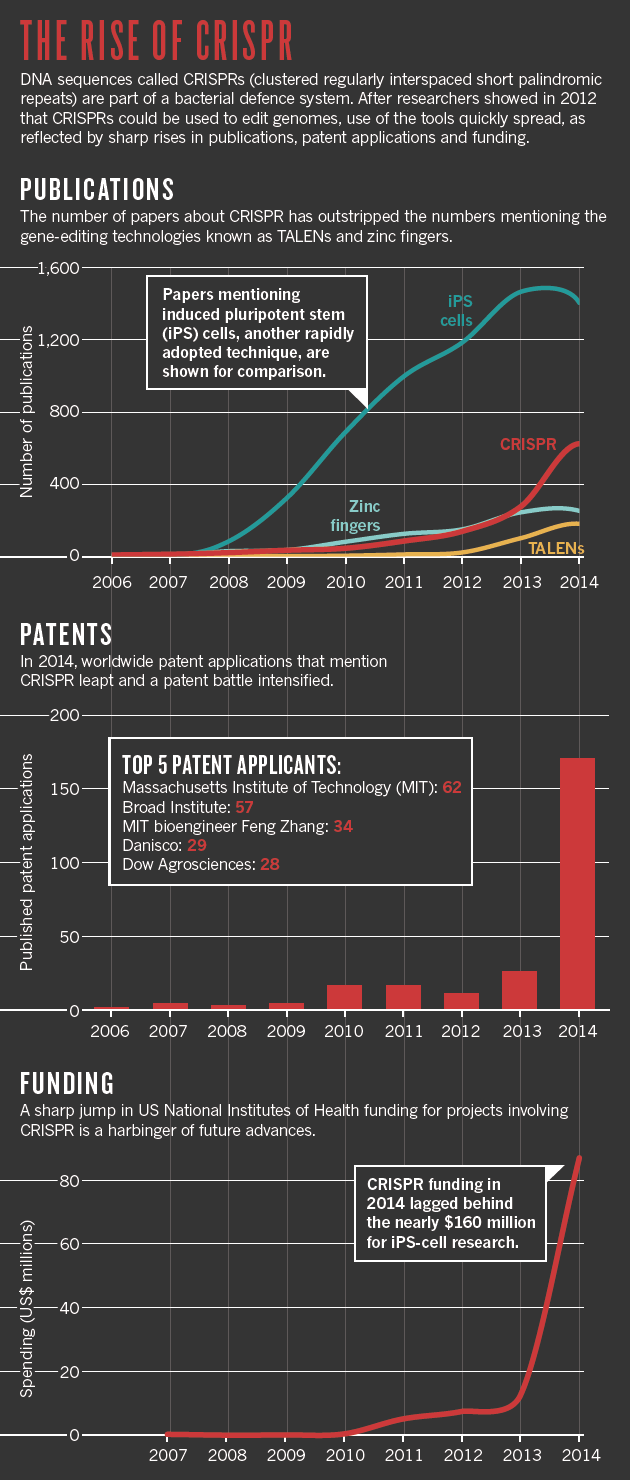
Publications: Scopus; Patents: The Lens; Funding: NIH RePORTER.
Researchers have traditionally relied heavily on model organisms such as mice and fruit flies, partly because they were the only species that came with a good tool kit for genetic manipulation. Now CRISPR is making it possible to edit genes in many more organisms. In April, for example, researchers at the Whitehead Institute for Biomedical Research in Cambridge, Massachusetts, reported using CRISPR to study Candida albicans, a fungus that is particularly deadly in people with weakened immune systems, but had been difficult to genetically manipulate in the lab3. Jennifer Doudna, a CRISPR pioneer at the University of California, Berkeley, is keeping a list of CRISPR-altered creatures. So far, she has three dozen entries, including disease-causing parasites called trypanosomes and yeasts used to make biofuels.
Yet the rapid progress has its drawbacks. “People just don’t have the time to characterize some of the very basic parameters of the system,” says Bo Huang, a biophysicist at the University of California, San Francisco. “There is a mentality that as long as it works, we don’t have to understand how or why it works.” That means that researchers occasionally run up against glitches. Huang and his lab struggled for two months to adapt CRISPR for use in imaging studies. He suspects that the delay would have been shorter had more been known about how to optimize the design of guide RNAs, a basic but important nuance.
By and large, researchers see these gaps as a minor price to pay for a powerful technique. But Doudna has begun to have more serious concerns about safety. Her worries began at a meeting in 2014, when she saw a postdoc present work in which a virus was engineered to carry the CRISPR components into mice. The mice breathed in the virus, allowing the CRISPR system to engineer mutations and create a model for human lung cancer4. Doudna got a chill; a minor mistake in the design of the guide RNA could result in a CRISPR that worked in human lungs as well. “It seemed incredibly scary that you might have students who were working with such a thing,” she says. “It’s important for people to appreciate what this technology can do.”
Andrea Ventura, a cancer researcher at Memorial Sloan Kettering Cancer Center in New York and a lead author of the work, says that his lab carefully considered the safety implications: the guide sequences were designed to target genome regions that were unique to mice, and the virus was disabled such that it could not replicate. He agrees that it is important to anticipate even remote risks. “The guides are not designed to cut the human genome, but you never know,” he says. “It’s not very likely, but it still needs to be considered.”
Last year, bioengineer Daniel Anderson of the Massachusetts Institute of Technology in Cambridge and his colleagues used CRISPR in mice to correct a mutation associated with a human metabolic disease called tyrosinaemia5. It was the first use of CRISPR to fix a disease-causing mutation in an adult animal — and an important step towards using the technology for gene therapy in humans (see ‘A brief history of CRISPR’).
The idea that CRISPR could accelerate the gene-therapy field is a major source of excitement in scientific and biotechnology circles. But as well as highlighting the potential, Anderson’s study showed how far there is to go. To deliver the Cas9 enzyme and its guide RNA into the target organ, the liver, the team had to pump large volumes of liquid into blood vessels — something that is not generally considered feasible in people. And the experiments corrected the disease-causing mutation in just 0.4% of the cells, which is not enough to have an impact on many diseases.
Over the past two years, a handful of companies have sprung up to develop CRISPR-based gene therapy, and Anderson and others say that the first clinical trials of such a treatment could happen in the next one or two years. Those first trials will probably be scenarios in which the CRISPR components can be injected directly into tissues, such as those in the eye, or in which cells can be removed from the body, engineered in the lab and then put back. For example, blood-forming stem cells might be corrected to treat conditions such as sickle-cell disease or β-thalassaemia. It will be a bigger challenge to deliver the enzyme and guide RNA into many other tissues, but researchers hope that the technique could one day be used to tackle a wider range of genetic diseases.

Yet many scientists caution that there is much to do before CRISPR can be deployed safely and efficiently. Scientists need to increase the efficiency of editing, but at the same time make sure that they do not introduce changes elsewhere in the genome that have consequences for health. “These enzymes will cut in places other than the places you have designed them to cut, and that has lots of implications,” says Haber. “If you’re going to replace somebody’s sickle-cell gene in a stem cell, you’re going to be asked, ‘Well, what other damage might you have done at other sites in the genome?’”
Keith Joung, who studies gene editing at Massachusetts General Hospital in Boston, has been developing methods to hunt down Cas9’s off-target cuts. He says that the frequency of such cuts varies widely from cell to cell and from one sequence to another: his lab and others have seen off-target sites with mutation frequencies ranging from 0.1% to more than 60%. Even low-frequency events could potentially be dangerous if they accelerate a cell’s growth and lead to cancer, he says.
With so many unanswered questions, it is important to keep expectations of CRISPR under control, says Katrine Bosley, chief executive of Editas, a company in Cambridge, Massachusetts, that is pursuing CRISPR-mediated gene therapy. Bosley is a veteran of commercializing new technologies, and says that usually the hard part is convincing others that an approach will work. “With CRISPR it’s almost the opposite,” she says. “There’s so much excitement and support, but we have to be realistic about what it takes to get there.”
While Anderson and others are aiming to modify DNA in human cells, others are targeting crops and livestock. Before the arrival of gene-editing techniques, this was generally done by inserting a gene into the genome at random positions, along with sequences from bacteria, viruses or other species that drive expression of the gene. But the process is inefficient, and it has always been fodder for critics who dislike the mixing of DNA from different species or worry that the insertion could interrupt other genes. What is more, getting genetically modified crops approved for use is so complex and expensive that most of those that have been modified are large commodity crops such as maize (corn) and soya beans.

Illustration by Sébastien Thibault
With CRISPR, the situation could change: the ease and low cost may make genome editing a viable option for smaller, speciality crops, as well as animals. In the past few years, researchers have used the method to engineer petite pigs and to make disease-resistant wheat and rice. They have also made progress towards engineering dehorned cattle, disease-resistant goats and vitamin-enriched sweet oranges. Doudna anticipates that her list of CRISPR-modified organisms will grow. “There’s an interesting opportunity to consider doing experiments or engineering pathways in plants that are not as important commercially but are very interesting from a research perspective — or for home vegetable gardens,” she says.
CRISPR’s ability to precisely edit existing DNA sequences makes for more-accurate modifications, but it also makes it more difficult for regulators and farmers to identify a modified organism once it has been released. “With gene editing, there’s no longer the ability to really track engineered products,” says Jennifer Kuzma, who studies science policy at North Carolina State University in Raleigh. “It will be hard to detect whether something has been mutated conventionally or genetically engineered.”
That rings alarm bells for opponents of genetically modified crops, and it poses difficult questions for countries trying to work out how to regulate gene-edited plants and animals. In the United States, the Food and Drug Administration has yet to approve any genetically modified animal for human consumption, and it has not yet announced how it will handle gene-edited animals.
Under existing rules, not all crops made by genome editing would require regulation by the US Department of Agriculture (see Nature 500, 389–390; 2013). But in May, the agriculture department began to seek input on how it can improve regulation of genetically modified crops — a move that many have taken as a sign that the agency is re-evaluating its rules in light of technologies such as CRISPR. “The window has been cracked,” says Kuzma. “What goes through the window remains to be seen. But the fact that it’s even been cracked is pretty exciting.”
Beyond the farm, researchers are considering how CRISPR could or should be deployed on organisms in the wild. Much of the attention has focused on a method called gene drive, which can quickly sweep an edited gene through a population. The work is at an early stage, but such a technique could be used to wipe out disease-carrying mosquitoes or ticks, eliminate invasive plants or eradicate herbicide resistance in pigweed, which plagues some US farmers.
Usually, a genetic change in one organism takes a long time to spread through a population. That is because a mutation carried on one of a pair of chromosomes is inherited by only half the offspring. But a gene drive allows a mutation made by CRISPR on one chromosome to copy itself to its partner in every generation, so that nearly all offspring will inherit the change. This means that it will speed through a population exponentially faster than normal (see ‘Gene drive’) — a mutation engineered into a mosquito could spread through a large population within a season. If that mutation reduced the number of offspring a mosquito produced, then the population could be wiped out, along with any malaria parasites it is carrying.
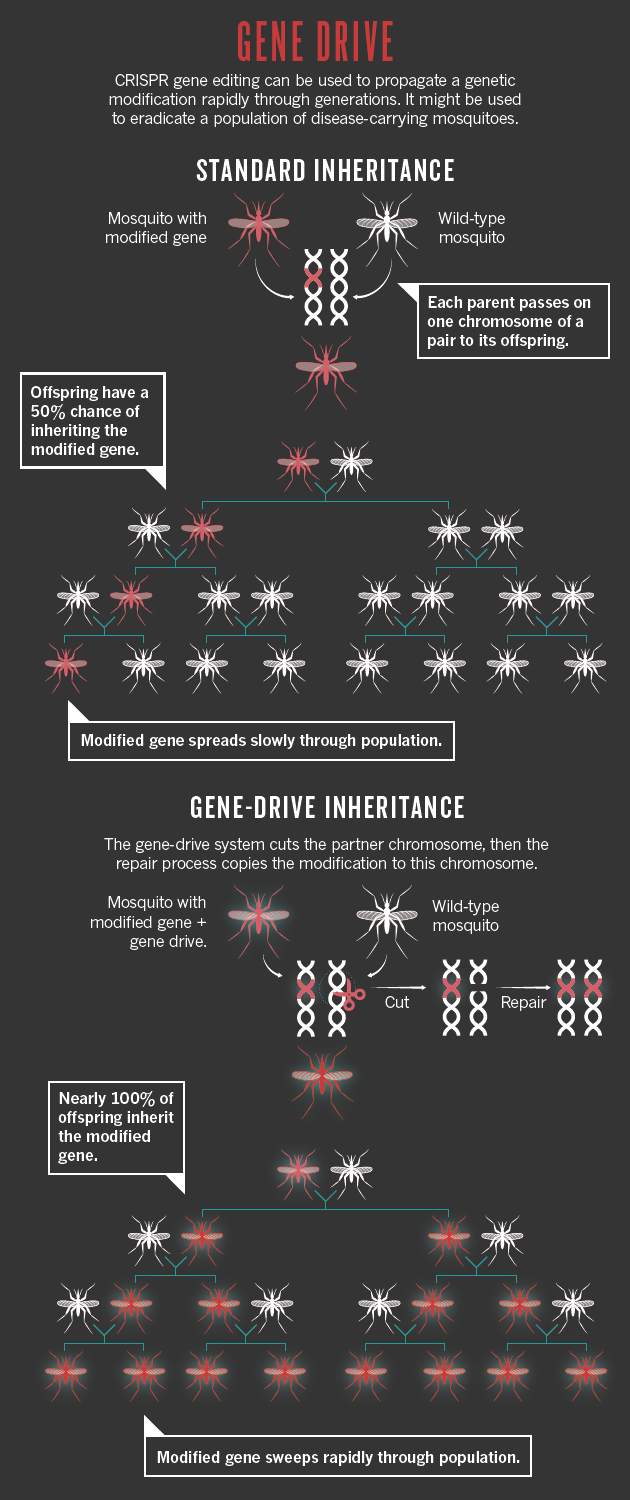
Publications: Scopus; Patents: The Lens; Funding: NIH RePORTER.
But many researchers are deeply worried that altering an entire population, or eliminating it altogether, could have drastic and unknown consequences for an ecosystem: it might mean that other pests emerge, for example, or it could affect predators higher up the food chain. And researchers are also mindful that a guide RNA could mutate over time such that it targets a different part of the genome. This mutation could then race through the population, with unpredictable effects.
“It has to have a fairly high pay-off, because it has a risk of irreversibility — and unintended or hard-to-calculate consequences for other species,” says George Church, a bioengineer at Harvard Medical School in Boston. In April 2014, Church and a team of scientists and policy experts wrote a commentary in Science6 warning researchers about the risks and proposing ways to guard against accidental release of experimental gene drives.
At the time, gene drives seemed a distant prospect. But less than a year later, developmental biologist Ethan Bier of the University of California, San Diego, and his student Valentino Gantz reported that they had designed just such a system in fruit flies7. Bier and Gantz had used three layers of boxes to contain their flies and adopted lab safety measures usually used for malaria-carrying mosquitoes. But they did not follow all the guidelines urged by the authors of the commentary, such as devising a method to reverse the engineered change. Bier says that they were conducting their first proof-of-principle experiments, and wanted to know whether the system worked at all before they made it more complex.
For Church and others, this was a clear warning that the democratization of genome editing through CRISPR could have unexpected and undesirable outcomes. “It is essential that national regulatory authorities and international organizations get on top of this — really get on top of it,” says Kenneth Oye, a political scientist at the Massachusetts Institute of Technology and lead author of the Science commentary. “We need more action.” The US National Research Council has formed a panel to discuss gene drives, and other high-level discussions are starting to take place. But Oye is concerned that the science is moving at lightning speed, and that regulatory changes may happen only after a high-profile gene-drive release.
The issue is not black and white. Micky Eubanks, an insect ecologist at Texas A&M University in College Station, says that the idea of gene drives shocked him at first. “My initial gut reaction was ‘Oh my god, this is terrible. It’s so scary’,” he says. “But when you give it more thought and weigh it against the environmental changes that we have already made and continue to make, it would be a drop in the ocean.”
Some researchers see lessons for CRISPR in the arc of other new technologies that prompted great excitement, concern and then disappointment when teething troubles hit. Medical geneticist James Wilson of the University of Pennsylvania in Philadelphia was at the centre of booming enthusiasm over gene therapy in the 1990s — only to witness its downfall when a clinical trial went wrong and killed a young man. The field went into a tailspin and has only recently begun to recover. The CRISPR field is still young, Wilson says, and it could be years before its potential is realized. “It’s in the exploration stage. These ideas need to ferment.”
Then again, Wilson has been bitten by the CRISPR bug. He says that he was sceptical of all the promises being made about it until his own lab began to play with the technique. “It’s ultimately going to have a role in human therapeutics,” he says. “It’s just really spectacular.”

a plant-hopping insect found in gardens across Europe – has hind-leg joints with curved cog-like strips of opposing ‘teeth’ that intermesh, rotating like mechanical gears to synchronise the animal’s legs when it launches into a jump.
The finding demonstrates that gear mechanisms previously thought to be solely man-made have an evolutionary precedent. Scientists say this is the “first observation of mechanical gearing in a biological structure“.
Through a combination of anatomical analysis and high-speed video capture of normal Issus movements, scientists from the University of Cambridge have been able to reveal these functioning natural gears for the first time. The findings are reported in the latest issue of the journal Science.
The gears in the Issus hind-leg bear remarkable engineering resemblance to those found on every bicycle and inside every car gear-box.
Each gear tooth has a rounded corner at the point it connects to the gear strip; a feature identical to man-made gears such as bike gears – essentially a shock-absorbing mechanism to stop teeth from shearing off.
The gear teeth on the opposing hind-legs lock together like those in a car gear-box, ensuring almost complete synchronicity in leg movement – the legs always move within 30 ‘microseconds‘ of each other, with one microsecond equal to a millionth of a second.
This is critical for the powerful jumps that are this insect’s primary mode of transport, as even miniscule discrepancies in synchronisation between the velocities of its legs at the point of propulsion would result in “yaw rotation” – causing the Issus to spin hopelessly out of control.
“This precise synchronisation would be impossible to achieve through a nervous system, as neural impulseswould take far too long for the extraordinarily tight coordination required,” said lead author Professor Malcolm Burrows, from Cambridge’s Department of Zoology.
“By developing mechanical gears, the Issus can just send nerve signals to its muscles to produce roughly the same amount of force – then if one leg starts to propel the jump the gears will interlock, creating absolutesynchronicity.
“We usually think of gears as something that we see in human designed machinery, but we’ve found that that is only because we didn’t look hard enough,” added co-author Gregory Sutton, now at the University of Bristol.
“These gears are not designed; they are evolved – representing high speed and precision machinery evolved for synchronisation in the animal world.”
Interestingly, the mechanistic gears are only found in the insect’s juvenile – or ‘nymph’ – stages, and are lost in the final transition to adulthood. These transitions, called ‘molts’, are when animals cast off rigid skin at key points in their development in order to grow.
It’s not yet known why the Issus loses its hind-leg gears on reaching adulthood. The scientists point out that a problem with any gear system is that if one tooth on the gear breaks, the effectiveness of the whole mechanism is damaged. While gear-teeth breakage in nymphs could be repaired in the next molt, any damage in adulthood remains permanent.
It may also be down to the larger size of adults and consequently their ‘trochantera’ – the insect equivalent of the femur or thigh bones. The bigger adult trochantera might allow them to can create enough friction to power the enormous leaps from leaf to leaf without the need for intermeshing gear teeth to drive it, say the scientists.
Each gear strip in the juvenile Issus was around 400 micrometres long and had between 10 to 12 teeth, with both sides of the gear in each leg containing the same number – giving a gearing ratio of 1:1.
Unlike man-made gears, each gear tooth is asymmetrical and curved towards the point where the cogs interlock – as man-made gears need a symmetric shape to work in both rotational directions, whereas the Issus gears are only powering one way to launch the animal forward.
While there are examples of apparently ornamental cogs in the animal kingdom – such as on the shell of the cog wheel turtle or the back of the wheel bug – gears with a functional role either remain elusive or have been rendered defunct by evolution.
The Issus is the first example of a natural cog mechanism with an observable function, say the scientists.
The Breakthrough Prize Foundation, an organization created by investor Yuri Milner, has announced that it will commit $100 million over the next ten years to a new, large-scale SETIinitiative.
 left – Greenbank observatory, West Virginia. right – Parkes observatory, Australia
left – Greenbank observatory, West Virginia. right – Parkes observatory, Australia
The intention is to use existing radio telescopes in West Virginia (the 100 meter Green Bank Telescope) and Australia (the 64 meter Parkes Telescope) to examine up to one million star systems for radio signals that would betray the presence of intelligence. The funding will allow the development of new receiving technologies that can speed up the search for radio broadcasts.
Beyond the nearby stellar targets, the project will also examine a large number of galaxies, as very advanced societies might be wielding high-powered signaling devices able to bridge intergalactic distances.
In addition to these radio SETI efforts, the Breakthrough Prize Foundation will underwrite an augmented search for laser flashes from the cosmos using a telescope at the Lick Observatory, in California. A further aspect of the initiative is to study the desirability and possible content of deliberate transmissions to nearby star systems, so-called active SETI.
Today’s announcement from the Breakthrough Prize Foundation is terrific news for all involved in SETI research. Private philanthropy plays an essential role in these efforts, and Yuri Milner’s contribution will fund SETI research on many fronts. The funding announced today does not go to the SETI Institute, but we are working with the Breakthrough Prize Foundation on other projects. The SETI Institute congratulates our colleagues and looks forward to joining them as additional Breakthrough Prize Foundation projects are rolled out. As ever, SETI Institute welcomes donations from all supporters of our work.
 image credit: breackthroughinitiatives.org
image credit: breackthroughinitiatives.org
The following commentary was published on July 20 in various media venues, and describes the societal motivation for SETI searches. It was signed by SETI Institute scientists Seth Shostak, Jill Tarter, and Emeritus Board Chairman, Frank Drake.
Who are we?
A mature civilization, like a mature individual, must ask itself this question. Is humanity defined by its divisions, its problems, its passing needs and trends? Or do we have a shared face, turned outward to the Universe?
In 1990, Voyager 1 swiveled its camera and captured the ‘Pale Blue Dot’ – an image of Earth from six billion kilometers away. It was a mirror held up to our planet – home of water, life, and minds. A reminder that we share something precious and rare.
But how rare, exactly? The only life? The only minds?
For the last half-century, small groups of scientists have listened valiantly for signs of life in the vast silence. But for government, academia, and industry, cosmic questions are astronomically far down the list of priorities. And that lengthens the odds of finding answers. It is hard enough to comb the Universe from the edge of the Milky Way; harder still from the edge of the public consciousness.
Yet millions are inspired by these ideas, whether they meet them in science or science fiction. Because the biggest questions of our existence are at stake. Are we the Universe’s only child – our thoughts its only thoughts? Or do we have cosmic siblings – an interstellar family of intelligence? As Arthur C. Clarke said, “In either case the idea is quite staggering.”
That means the search for life is the ultimate ‘win-win’ endeavor. All we have to do is take part.
Today we have search tools far surpassing those of previous generations. Telescopes can pick out planets across thousands of light years. The magic of Moore’s law lets our computers sift data orders of magnitude faster than older mainframes – and ever quicker each year.
These tools are now reaping a harvest of discoveries. In the last few years, astronomers and the Kepler Mission have discovered thousands of planets beyond our solar system. It now appears that most stars host a planetary system. Many of them have a planet similar in size to our own, basking in the ‘habitable zone’ where the temperature permits liquid water. There are likely billions of earth-like worlds in our galaxy alone. And with instruments now or soon available, we have a chance of finding out if any of these planets are true Pale Blue Dots – home to water, life, even minds.
There has never been a better moment for a large-scale international effort to find life in the Universe. As a civilization, we owe it to ourselves to commit time, resources, and passion to this quest.
But as well as a call to action, this is a call to thought. When we find the nearest exo-Earth, should we send a probe? Do we try to make contact with advanced civilizations? Who decides? Individuals, institutions, corporations, or states? Or can we as species – as a planet – think together?
Three years ago, Voyager 1 broke the sun’s embrace and entered interstellar space. The 20th century will be remembered for our travels within the solar system. With cooperation and commitment, the present century will be the time when we graduate to the galactic scale, seek other forms of life, and so know more deeply who we are.