Category: Uncategorized
Vi.Tri_Ne Sub:JECti:Ve
Durante la próxima edición de la Nuit Blanche, el artista Arcángelo Constantini en colaboración con Rodrigo Sigal, Iracema de Andrade, Skot Deeming y Jorge Ramírez, presentarán bakteria.org Vi.Tri_NA Sub:JEti:Va. El proyecto multidisciplinario se basa en la improvisación y la presentación de procesos creativos en tiempo real, así como en la experimentación con nuevos medios y nuevas tecnologías.
Pour la prochaine edition de la Nuit Blanche, l’artist Arcangelo Constantini en collaboration avec Rodrigo Sigal, Iracema de Andrade, Skot Deeming et Jorge Ramírez, vous présenteront bakteria.org Vi.Tri_Ne Sub:JECti:Ve. Ce projet multidisciplinaire est basé sur improvisation et la présentation de processus de création en temps réel, ainsi que sur expérimentation de nouveaux médias et de nouvelles technologies.
For the next edition of the Nuit Blanche, the artist Arcangelo Constantini in collaboration with Rodrigo Sigal, Iracema de Andrade, Skot Deeming and Jorge Ramírez, will be presenting bakteria.org Sub:JECti:Ve Vi.Tri_Ne. The multidisciplinary project is based on improvisation and the presentation of creative processes in real time, as well as experimentation with new media and new techologies.
PET PLàsTïK. PöLLuTiöN. mOLE,KUL.àR. bre!AKER.

http://www.theguardian.com/environment/2016/mar/10/could-a-new-plastic-eating-bacteria-help-combat-this-pollution-scourge
Nature has begun to fight back against the vast piles of filth dumped into its soils, rivers and oceans by evolving a plastic-eating bacteria – the first known to science.
In a report published in the journal Science, a team of Japanese researchers described a species of bacteria that can break the molecular bonds of one of the world’s most-used plastics – polyethylene terephthalate, also known as PET or polyester.
The Japanese research team sifted through hundreds of samples of PET pollution before finding a colony of organisms using the plastic as a food source.
Further tests found the bacteria almost completely degraded low-quality plastic within six weeks. This was voracious when compared to other biological agents; including a related bacteria, leaf compost and a fungus enzyme recently found to have an appetite for PET.
“This is the first rigorous study – it appears to be very carefully done – that I have seen that shows plastic being hydrolyzed [broken down] by bacteria,” said Dr Tracy Mincer, a researcher at Woods Hole Oceanographic Institution.
The molecules that form PET are bonded very strongly, said Prof Uwe Bornscheuer in an accompanying comment piece in Science. “Until recently, no organisms were known to be able to decompose it.”
In a Gaian twist, initial genetic examination revealed the bacteria, namedIdeonella sakaiensis 201-F6, may have evolved enzymes specifically capable of breaking down PET in response to the accumulation of the plastic in the environment in the past 70 years.
Such rapid evolution was possible, said Enzo Palombo, a professor of microbiology at Swinburne University, given that microbes have an extraordinary ability to adapt to their surroundings. “If you put a bacteria in a situation where they’ve only got one food source to consume, over time they will adapt to do that,” he said.
“I think we are seeing how nature can surprise us and in the end the resiliency of nature itself,” added Mincer.
The bacteria took longer to eat away highly crystallised PET, which is used in plastic bottles. That means the enzymes and processes would need refinement before they could be useful for industrial recycling or pollution clean-up.
“It’s difficult to break down highly crystallised PET,” said Prof Kenji Miyamoto from Keio University, one of the authors of the study. “Our research results are just the initiation for the application. We have to work on so many issues needed for various applications. It takes a long time,” he said.
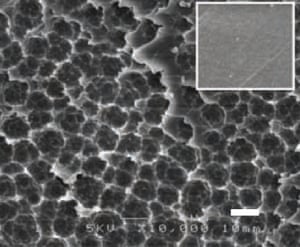
A third of all plastics end up in the environment and 8m tonnes end up in the ocean every year, creating vast accumulations of life-choking rubbish.
PET makes up almost one-sixth of the world’s annual plastic production of 311m tons. Despite PET being one of the more commonly recycled plastics, the World Economic Forum (WEF) reports that only just over half is ever collected for recycling and far less actually ends up being reused.
Advances in biodegradable plastics and recycling offer hope for the future, said Bornscheuer, “but [this] does not help to get rid of the plastics already in the environment”.
However the potential applications of the discovery remain unclear. The most obvious use would be as a biological agent in nature, said Palombo. Bacteria could be sprayed on the huge floating trash heaps building up in the oceans. This method is most notably employed to combat oil spills.
This particular bacteria would not be useful for this process as it only consumes PET, which is too dense to float on water. But Bornscheuer said the discovery could open the door to the discovery or manufacture of biological agents able to break down other plastics.
Palombo said the discovery suggested that other bacteria may have already evolved to do this job and simply needed to be found.
“I would not be surprised if samples of ocean plastics contained microbes that are happily growing on this material and could be isolated in the same manner,” he said.
But Mincer said breaking down ocean rubbish came with dangers of its own.Plastics often contain additives that can be toxic when released. WEF estimates that the 150m tonnes of plastic currently in the ocean contain roughly 23m tonnes of additives.
“Plastic debris may have been less toxic in the whole unhydrolyzed form where it would ultimately have been buried in the sediments on a geological timescale,” said Mincer.
Beyond dealing with the plastic already fouling up the environment, the bacteria could potentially be used in industrial recycling processes.
“Certainly, the use of these microbes or enzymes could play a role in remediation of plastic in a controlled reactor,” said Mincer.
Miyamoto’s team suggested that the environmentally-benign constituents left behind by the bacteria could be the same ones from which the plastic is formed. If this were true and a process could be developed to isolate them, Bornscheuer said: “This could provide huge savings in the production of new polymer without the need for petrol-based starting materials.” According to the WEF, 6% of global oil production is devoted to the production of plastics.
But the plastics industry said the potential for a new biological process to replace or augment the current mechanical recycling process was very small.
“PET is 100% recyclable,” said Mike Neal, the chairman of the Committee of PET Manufacturers in Europe. “I expect that a biodegradation system would require a similar engineering process to chemical depolymerisation and as such is unlikely to be economically viable,” he said.
AKziön Vi-SuaL:So-NiKa
Vi3R-NEz 27 , MaYo 20:00
MüSéö de ArTe Con-Tem-PoRa:NeÒ OãXãCã
AKziön Vi-SuaL:So-NiKa

El Prin-CiPîö de La FöRMa ((( 3VôLûCîôN-Muta-CîöN ))) Sci:EnTi-StS un.VeiL :N3W Tr33 Of Li.Feee
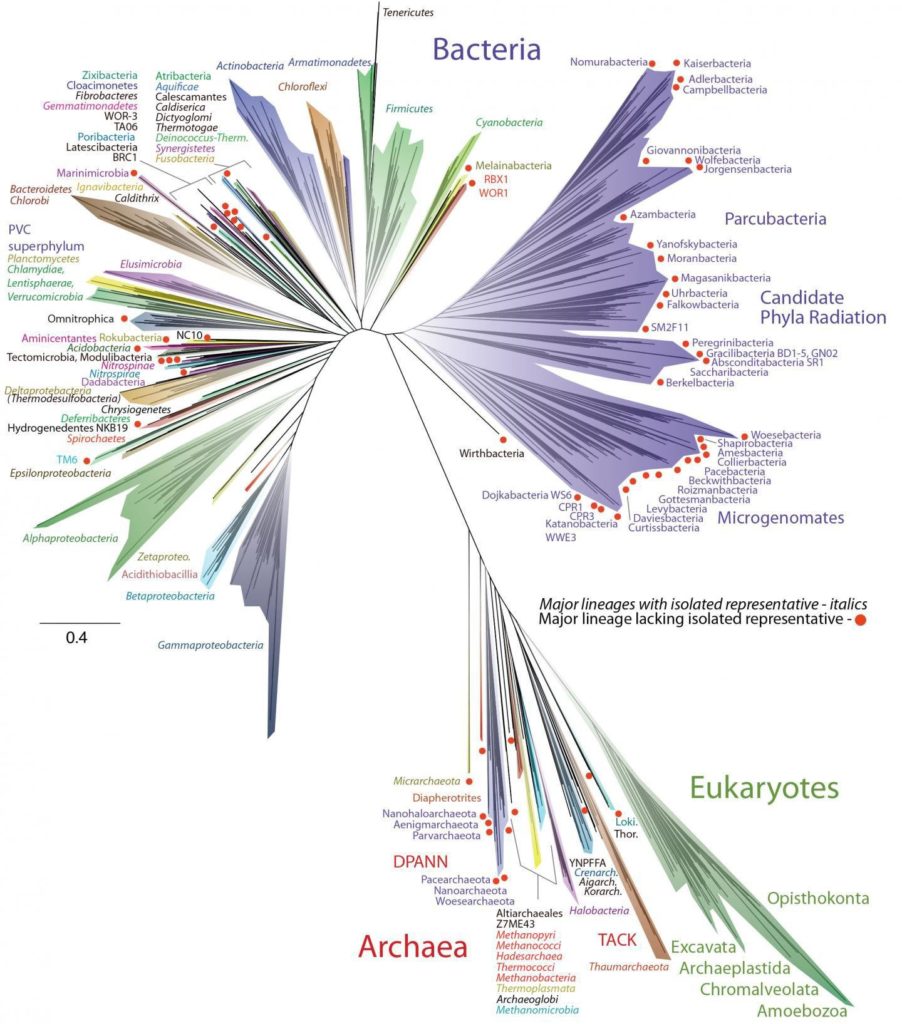
A team of scientists unveiled a new tree of life on Monday, a diagram outlining the evolution of all living things. The researchers found that bacteria make up most of life’s branches. And they found that much of that diversity has been waiting in plain sight to be discovered, dwelling in river mud and meadow soils.
“It is a momentous discovery — an entire continent of life-forms,” said Eugene V. Koonin of the National Center for Biotechnology Information, who was not involved in the study.
The study was published in the journal Nature Microbiology.
In his 1859 book “On the Origin of Species,” Charles Darwin envisioned evolution like a branching tree. The “great Tree of Life,” he said, “fills with its dead and broken branches the crust of the earth, and covers the surface with its ever branching and beautiful ramifications.”
Ever since, biologists have sought to draw the tree of life. The invention of DNA sequencing revolutionized that project, because scientists could find the relationship among species encoded in their genes.
In the 1970s, Carl Woese of the University of Illinois and his colleagues published the first “universal tree of life” based on this approach. They presented the tree as three great trunks.
Our own trunk, known as eukaryotes, includes animals, plants, fungi and protozoans. A second trunk included many familiar bacteria like Escherichia coli.
The third trunk that Woese and his colleagues identified included little-known microbes that live in extreme places like hot springs and oxygen-free wetlands. Woese and his colleagues called this third trunk Archaea.

Scientists who wanted to add new species to this tree of life have faced a daunting challenge: They do not know how to grow the vast majority of single-celled organisms in their laboratories.
A number of researchers have developed a way to get around that. They simply pull pieces of DNA out of the environment and piece them together.
In recent years, Jillian F. Banfield of the University of California, Berkeley and her colleagues have been gathering DNA from many environments, like California meadows and deep sea vents. They have been assembling the genomes of hundreds of new microbial species.
The scientists were so busy reconstructing the new genomes that they did not know how these species might fit on the tree of life. “We never really put the whole thing together,” Dr. Banfield said.
Recently, Dr. Banfield and her colleagues decided it was time to redraw the tree.
They selected more than 3,000 species to study, bringing together a representative sample of life’s diversity. “We wanted to be as comprehensive as possible,” said Laura A. Hug, an author of the new study and a biologist at the University of Waterloo in Canada.
The researchers studied DNA from 2,072 known species, along with the DNA from 1,011 species newly discovered by Dr. Banfield and her colleagues.
The scientists needed a supercomputer to evaluate a vast number of possible trees. Eventually, they found one best supported by the evidence.
It’s a humbling thing to behold. All the eukaryotes, from humans to flowers to amoebae, fit on a slender twig. The new study supported previous findings that eukaryotes and archaea are closely related. But overshadowing those lineages is a sprawling menagerie of bacteria.
Remarkably, the scientists didn’t have to go to extreme places to find many of their new lineages. “Meadow soil is one of the most microbially complex environments on the planet,” Dr. Hug said.
Another new feature of the tree is a single, large branch that splits off near the base. The bacteria in this group tend to be small in size and have a simple metabolism.
Dr. Banfield speculated that they got their start as simple life-forms in the first chapters in the history of life. They have stuck with that winning formula ever since.
“This is maybe an early evolving group,” Dr. Banfield said. “Their advantage is just being around for a really long time.”
Brian P. Hedlund, a microbiologist at the University of Nevada, Las Vegas who was not involved in the new study, said that one of the most striking results of the study was that the tree of life was dominated by species that scientists have never been able to see or grow in their labs. “Most of life is hiding under our noses,” he said.
Patrick Forterre, an evolutionary biologist at the Pasteur Institute in France, agreed that bacteria probably make up much of life’s diversity. But he had concerns about how Dr. Banfield and her colleague built their tree. He argued that genomes assembled from DNA fragments could actually be chimeras, made up of genes from different species. “It’s a real problem,” he said.
Dr. Banfield predicted that the bacterial branches of the tree of life may not change much in years to come. “We’re starting to see the same things over and over again,” she said.
Instead, Dr. Banfield said she expected new branches to be discovered for eukaryotes, especially for tiny species such as microscopic fungi. “That’s where I think the next big advance might be found,” Dr. Banfield said.
Dr. Hug disagreed that scientists were done with bacteria. “I’m less convinced we’re hitting a plateau,” she said. “There are a lot of environments still to survey.”
A picture caption on Tuesday with an article about a new tree of life published by scientists referred incorrectly to Methanosarcina, the organism shown. It belongs to the domain archaea, not bacteria.
QuanTuM, (tELe)TRaNS:pOR:TATion Of Micro:OrgA,NisimS
So:uR_Ce : http://phys.org/news/2016-01-physicists-scheme-teleport-memory.html
Physicists propose the first scheme to teleport the memory of an organism
January 14, 2016
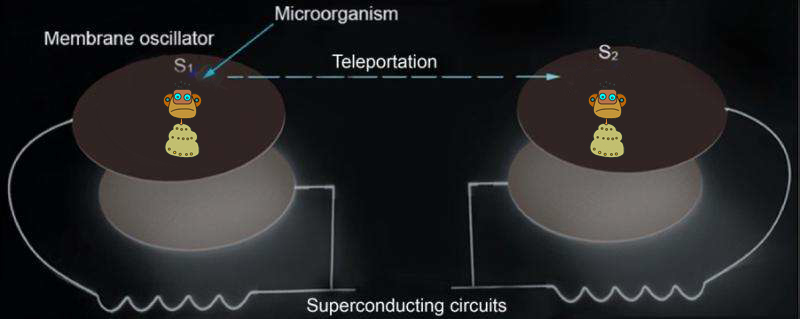
In “Star Trek,” a transporter can teleport a person from one location to a remote location without actually making the journey along the way. Such a transporter has fascinated many people. Quantum teleportation shares several features of the transporter and is one of the most important protocols in quantum information. In a recent study, Prof. Tongcang Li at Purdue University and Dr. Zhang-qi Yin at Tsinghua University proposed the first scheme to use electromechanical oscillators and superconducting circuits to teleport the internal quantum state (memory) and center-of-mass motion state of a microorganism. They also proposed a scheme to create a Schrödinger’s cat state in which a microorganism can be in two places at the same time. This is an important step toward potentially teleporting an organism in future.
In 1935, Erwin Schrödinger proposed a famous thought experiment to prepare a cat in a superposition of both alive and dead states. The possibility of an organism to be in a superposition state dramatically reveals the profound consequences of quantum mechanics, and has attracted broad interests. Physicists have made great efforts over many decades to investigate macroscopic quantum phenomena. To date, matter-wave interference of electrons, atoms, and molecules (such as C60) have been observed. Recently, quantum ground state cooling and the creation of superposition states of mechanical oscillators have been realized. For example, a group in Colorado, U.S. has cooled the vibration of a 15-micrometer-diameter aluminum membrane to quantum ground state, and entangled its motion with microwave photons. However, the quantum superposition of an entire organism has not been realized. Meanwhile, there have been many breakthroughs in quantum teleportationsince its first experimental realization in 1997 with a single photon. Besides photons, quantum teleportation with atoms, ions, and superconducting circuits have been demonstrated. In 2015, a group at University of Science and Technology of China demonstrated the quantum teleportation of multiple degrees of freedom of a single photon. However, existing experiments are still far away from teleporting an organism or the state of an organism.
In a recent study, Tongcang Li and Zhang-qi Yin propose to put a bacterium on top of an electromechanical membrane oscillator integrated with a superconducting circuit to prepare the quantum superposition state of a microorganism and teleport its quantum state. A microorganism with a mass much smaller than the mass of the electromechanical membrane will not significantly affect the quality factor of the membrane and can be cooled to the quantum ground state together with the membrane. Quantum superposition and teleportation of its center-of-mass motion state can be realized with the help of superconducting microwave circuits. With a strong magnetic field gradient, the internal states of a microorganism, such as the electron spin of a glycine radical, can be entangled with its center-of-mass motion and be teleported to a remote microorganism. Since internal states of an organism contain information, this proposal provides a scheme for teleporting information or memories between two remote organisms.
The proposed setup is also a quantum-limited magnetic resonance force microscope. It can not only detect the existence of single electron spins (associated with protein defects or DNA defects) like conventional MRFM, but can also coherently manipulate and detect the quantum states of electron spins. It enables some isolated electron spins that could not be read out with optical or electrical methods to be used as quantum memory for quantum information.
Li says, “We propose a straightforward method to put a microorganism in two places at the same time, and provide a scheme to teleport the quantum state of a microorganism. I hope our unconventional work will inspire more people to think seriously about quantum teleportation of a microorganism and its potential applications in the future.” Yin says “Our work also provides insights for future studies about the effects of biochemical reactions in the wave function collapses of quantum superposition states of an organism.”
Read more at: http://phys.org/news/2016-01-physicists-scheme-teleport-memory.html#jCp
Ci;ENTiFi;Koz Vou:YE;Ri.Ztaz , VieN:Do PoRNO Eu:KA-RyO_TiKO El SeX.O en :LoS., Or;GA;NIzMoz _ MuL:Ti.Ce.Lu*LA.Rez
sou:RCe :
Sex among eukaryotes is far more common than once believed
July 28, 2015 by Christopher Packham report
(Phys.org)—For a long time, biologists have considered sex to be an inherent trait of multicellular life, while microbial eukaryotes were considered to be either optionally sexual or purely clonal. From this perspective, clonality in eukaryotes is seen as exceptional. However, a group of researchers Europe and Canada have recently published a paper examining this broad distinction more closely, and have suggested that it appears to stem from an improper comparison of unicellular and multicellular species.
The paper, published in the Proceedings of the National Academy of Sciences, points out that sex in multicellular organisms is simply clonal cell propagation among physically linked cells. “Hence,” the researchers write, “from the perspective of cell lineage, sex in multicellular organisms is as episodic as it is in facultatively sexual unicellular eukaryotes.” The authors’ emphatic conclusion is that “sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life.”
Notably, the paper emphasizes that zoologists would be aghast at the absence of observed sex, while microbiologists are far more receptive to the lack of sex in protists. Many protist groups, including ciliates and green algae, propagate via sex, but direct observation of those processes is lacking for the vast majority. Indeed, there are entire lineages of protists for which no evidence of sex processes exists. However, the authors screened scientific literature to find individual “signs of sex” in eukaryotic lineages, including physical observation of cell fusion or nuclear fusion, genetic evidence of meiosis or recombination, or changes in ploidy levels over the life cycle.
Among the individuals screened, Jakobida, Glaucophyta, and Malawaimonadida—putatively asexual eukaryotes— were all found to contain genes involved in gamete fusion and/or nuclear fusion. The authors suggest that sex among unicellular eukaryotes is likely to be far more common than currently believed, and the lack of evidence of sexual propagation attributable to the difficulty of microbiological observation. Highlighting this difficulty, they point out a famous example of a particular type of algae with two morphologically different life cycle stages, which had been wrongly considered to be two separate species. What we don’t know about protist life forms still vastly outweighs what we’ve discovered. “…(W)e still have a tendency to underestimate how widespread sexual practices are in the different eukaryotic groups,” the authors write.
Further, genome sequencing now supports the fundamentally sexual nature of eukaryotes. The authors cite numerous examples of putatively asexual eukaryotes found to express genetic traits associated with sex propagation. Giardia intestinalis was assumed to be asexual until genomic inspection revealed allelic differences indicative of sex. “The list of eukaryotic species that lack strong direct evidence for meiotic sex, but that seem sexual, as suggested by the presence of these meiosis-associated genes, is growing longer and longer,” the authors write.
There are numerous adaptive benefits to sex: It creates genetic variation, repairs DNA breaks, and prevents the accumulation of disadvantageous mutations. Sexual reproduction is also associated with species survival during adverse periods. Because of these advantages, the authors suggest, even asexual species overwhelmingly retain the option for meiotic sex propagation, even despite some of the disadvantages of sex for protists.
The paper goes on to speculate on the possibility that the evolution of meiotic sex was a defensive response to DNA-damaging effects of reactive oxygen species, and considers the possible influence of endosymbiotic organisms like chloroplasts and mitochondria on the evolution of sex.
![]() Explore further: Researchers Present New Sex Evolution Theory
Explore further: Researchers Present New Sex Evolution Theory
More information: “Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life.” PNAS 2015 112 (29) 8827-8834; published ahead of print July 21, 2015, DOI: 10.1073/pnas.1501725112
Abstract
Sexual reproduction and clonality in eukaryotes are mostly seen as exclusive, the latter being rather exceptional. This view might be biased by focusing almost exclusively on metazoans. We analyze and discuss reproduction in the context of extant eukaryotic diversity, paying special attention to protists. We present results of phylogenetically extended searches for homologs of two proteins functioning in cell and nuclear fusion, respectively (HAP2 and GEX1), providing indirect evidence for these processes in several eukaryotic lineages where sex has not been observed yet. We argue that (i) the debate on the relative significance of sex and clonality in eukaryotes is confounded by not appropriately distinguishing multicellular and unicellular organisms; (ii) eukaryotic sex is extremely widespread and already present in the last eukaryotic common ancestor; and (iii) the general mode of existence of eukaryotes is best described by clonally propagating cell lines with episodic sex triggered by external or internal clues. However, important questions concern the relative longevity of true clonal species (i.e., species not able to return to sexual procreation anymore). Long-lived clonal species seem strikingly rare. We analyze their properties in the light of meiotic sex development from existing prokaryotic repair mechanisms. Based on these considerations, we speculate that eukaryotic sex likely developed as a cellular survival strategy, possibly in the context of internal reactive oxygen species stress generated by a (proto) mitochondrion. Thus, in the context of the symbiogenic model of eukaryotic origin, sex might directly result from the very evolutionary mode by which eukaryotic cells arose.
Read more at: http://phys.org/news/2015-07-sex-eukaryotes-common-believed.html#jCp
So:Me i,NNo:Cu.OuS Mi,Cro.OrgANiss:Ms W.E CaRR-Y Can,. A.FFE.Kt Be.HA_VioR . AnD, . Cog.Ni:ti.On
Sou:RCe
Algae in Your Throat? Scientists Discover Algae Virus in Humans
FAST FACTS:
- Scientists have discovered that some healthy people carry in their throats a green algae virus previously thought to be non-infectious to humans.
- The virus may cause subtle cognitive changes in some.
- The study highlights the potential of otherwise innocuous organisms to affect physiologic functions without causing outright disease.

Scientists from Johns Hopkins and the University of Nebraska have discovered an algae virus never before seen in the throats of healthy people that may subtly alter a range of cognitive functions including visual processing and spatial orientation in those who harbor it. A report on the team’s findings is published online Oct. 27 in Proceedings of the National Academy of Science.
The discovery casts in a new light a class of viruses that has been thus far deemed non-infectious to humans, underscoring the ability of certain microorganisms to trigger delicate physiologic changes without causing full-blown disease, the researchers say.
“This is a striking example showing that the ‘innocuous’ microorganisms we carry can affect behavior and cognition,” says lead investigator Robert Yolken, M.D., a virologist and pediatric infectious disease specialist at the Johns Hopkins Children’s Center and director of the Stanley Neurovirology Laboratory at Johns Hopkins. “Many physiological differences between person A and person B are encoded in the set of genes each inherits from parents, yet some of these differences are fueled by the various microorganisms we harbor and the way they interact with our genes.”
People’s bodies are colonized by trillions of bacteria, viruses and fungi, a constellation of organisms whose functions are largely unknown and collectively make up the so-called human microbiome. Many are presumed harmless, while others, such asLactobacillus acidophilus, are known to have clear benefits for human health. The findings of the new research suggest some may also affect human health in less obvious and not entirely benign ways.
In addition, the study provides a rare proof of a biologic phenomenon known as viral jumping, which typically occurs when viruses cross over from one species to another — think avian and swine flu — but is rarely seen across biologic kingdoms.
Yolken and colleagues stumbled upon the algae virus unexpectedly while analyzing the microbial population of the throats of healthy humans for a non-related study. Investigators obtained throat swabs and performed DNA analysis designed to detect the genetic footprints of viruses and bacteria. To their surprise, the researchers say, they discovered DNA matching that of Acanthocystis turfacea Chlorella virus 1, or ATCV-1, known to infect green algae. Green algae include more than 7,000 water-dwelling organisms that resemble plants but belong to a separate biologic kingdom. They are commonly found in aquatic environments like ponds, lakes and the ocean.
Forty of 92 participants in the study tested positive for the algae virus. The group that harbored the virus performed worse overall on a set of tasks to measure the speed and accuracy of visual processing. While their performance was not drastically poorer, it was measurably lower, the researchers say. For example, people who harbored the virus scored, on average, nearly nine points lower on a test that measured how quickly they could draw a line between sequentially numbered circles on a piece of paper. Viral carriers also scored seven points lower, on average, on tests measuring attention.
To further elucidate the effects of the virus, the investigators infected a group of mice and analyzed their performance on a set of tests designed to measure the rodent equivalent of human cognitive function. Animals infected with the virus exhibited deficits similar to those observed in humans. Infected animals had worse recognition memory and spatial orientation than uninfected mice. For example, they had a harder time finding their way around a maze, failing to recognize a new entry that was previously inaccessible. In addition, infected animals were less likely to pay attention to a new object, spending nearly 30 percent less time exploring it than uninfected mice, a finding that suggest shorter attention span and greater distractibility. The researchers caution that drawing direct links between mice and humans can be reductive but, they say, the parallels observed in the study were rather striking.
“The similarity of our findings in mice and humans underscores the common mechanisms that many microbes use to affect cognitive function in both animals and people,” says co-investigator Mikhail Pletnikov, M.D., Ph.D., director of the Behavioral Neurobiology and Neuroimmunology Laboratory at Johns Hopkins. “This commonality is precisely what allows us to study the pathologies that these microorganisms fuel and do so in a controlled systematic way.”
Analysis of brain samples from virus-infected mice revealed changes in the expression of multiple genes found in the hippocampus, the part of the brain that sorts and catalogues short-term and long-term memories and guides spatial orientation. Some of these alterations involved genes that regulate brain response to dopamine — a neurotransmitter affecting a wide range of neurologic and cognitive functions — as well as genes involved in immune cell regulation. The finding of multiple gene involvement suggests numeous mechanisms that may explain some of the effects observed in the study, the researchers say. The investigators, however, caution that their findings require in-depth follow-up to clarify the effects of the virus on human cognition and the exact mechanisms that precipitate them.
The new findings come on the heels of several recent studies showing that microbes can play an important role in the genesis of neurologic, cognitive and mental health disorders. For example, Yolken and others have previously shown that infection with the cat-borne parasite Toxoplasma gondii can alter behavior in genetically predisposed people — an illustrative example of the often synergistic role that genes and environment can play in human disease.
James Van Etten of the University of Nebraska is an expert on algal viruses and was senior author on the paper. Other researchers from the University of Nebraska included David Dunigan, James Gurnon, Fangrui Ma and Irina Agarkova.
Other Johns Hopkins investigators included Lorraine Jones-Brando, Geetha Kannan, Emily Severance, Sarven Subunciyan, C. Conover Talbot Jr., Emese Prandovszky, Flora Leister, Kristen Gressitt, Ou Chen and Bryan Deuber.
Faith Dickerson of Sheppard Pratt Health System in Baltimore was also a co-investigator.
The research was funded by the Stanley Medical Research Institute, the National Science Foundation and the National Center for Research Resources, part of the National Institutes of Health, under grant number P20-RR15635.
A Ge:Ne:Tik . Swi:TCh To , StoP A,Ging De,GenE:Ra:TiiNg CeLLs Un I,NTe;RruPTor , GeNe;Tiko , Pa:RA De:Te:NEr , El En:;VeJe:Zi Mi: ento ( y) La De;Ge;NEraCiiOn , CeLU:LAr
A new study, conducted by researchers from Northwestern University in Illinois, has revealed that a simple genetic “switch” may be the key to the solving the mystery of aging.
The study of worms showed adult cells abruptly begin their downhill slide when they reach reproductive maturity. A genetic switch then allows aging to begin by “turning off” certain processes which protect cells within the body.
The finding is significant because humans have the same genetic switch – and means eventually it may be possible to delay aging and certain degenerative diseases.
Genetic switches then start the aging process by turning off cell stress responses that protect cells by keeping important proteins folded and functional. The results, published in the journal Molecular Cell, claim to pinpoint the start of aging, disproving the theory that aging is a slow series of random events.
Researchers studied the transparent roundworm C. elegans, and found this “switch” is thrown by germline stem cells in early adulthood after it starts to reproduce ensuring its line will live on. C. elegans have a biochemical environment similar to that of humans and are a popular research tool for the study of the biology of aging and are used to model human diseases. Knowing more about how the quality control system works in cells could help researchers one day figure out how to delay degenerative diseases related to aging, such as neuro-degenerative diseases.
“Wouldn’t it be better for society if people could be healthy and productive for a longer period during their lifetime?” said Richard Morimoto, the senior author of the study said.
“I am very interested in keeping the quality control systems optimal as long as we can, and now we have a target. Our findings suggest there should be a way to turn this genetic switch back on and protect our aging cells by increasing their ability to resist stress.” The scientists found in C. elegans the decline begins eight hours into adulthood, when all of the switches get thrown to shut off the animal’s cell stress protective mechanisms. Professor Morimoto also found it is the germline stem cells responsible for making eggs and sperm that controls the switch.
In animals including C. elegans and humans the heat shock response is essential for proper protein folding and cellular health. Aging is associated with a decline in quality control, so Morimoto looked specifically at the heat shock response in the life of C. elegans. “We saw a dramatic collapse of the protective heat shock response beginning in early adulthood,” he said. “C. elegans has told us that aging is not a continuum of various events, which a lot of people thought it was. In a system where we can actually do the experiments, we discover a switch that is very precise for aging. All these stress pathways that insure robustness of tissue function are essential for life, so it was unexpected that a genetic switch is literally thrown eight hours into adulthood, leading to the simultaneous repression of the heat shock response and other cell stress responses.”
Using a combination of genetic and biochemical approaches, Professor Morimoto found the protective heat shock response declines steeply over a four-hour period in early adulthood, precisely at the onset of reproductive maturity. The animals still appear normal in behaviour, but the scientists can see molecular changes and the decline of protein quality control. In one experiment, the researchers blocked the germline from sending the signal to turn off cellular quality control. They found the somatic tissues remained robust and stress resistant in the adult animals.
Er:NsZt H(ae)ck3L
Ar;T ForMs Of NAtU:Re // BuZk:Ando InS,PiRa:Zion En La EZ,Pe;Ku,LAZion , EZtETi;KA de _ Er:NsZt H(ae)ck3L .pa:Ra , Ini:CiAr, La : In:TeNSa LAbOr Mor:PHo.LOGi;KA de bakteria.org Kla:;Si.Fi:KArr CoNTe:Xtos ,Com;PArAti;Voz En La Ge;NeSiS D(e) Las , ra_MIFi;KA:ci_Onez Kre:Azio:NizTAz Ez.PonTa:Ne:az